Downloading from BaseSpace
Dave Tang
2019-07-12
Last updated: 2019-07-12
Checks: 6 0
Knit directory: listerlab/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190712) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0051254 | davetang | 2019-07-12 | Removed password |
| html | a0ef9b7 | davetang | 2019-07-12 | Build site. |
| Rmd | 389bd42 | davetang | 2019-07-12 | First commit |
Introduction
This guide provides directions for downloading raw sequencing data from BaseSpace. The BaseSpace Sequence Hub is a cloud-based genomics analysis and storage platform that directly integrates with all Illumina sequencers.
Connecting to the server
SSH into Razor and start or resume a screen session.
ssh -Y -i your_private_key -p 2020 your_username@202.8.39.31
# name your screen session name as basemount
screen -S basemountCreating a new directory
Sequencing data is stored on a network mount: /mnt/remoteserv/switch/rundata. Firstly we need to create a new directory for storing our data. The following nomenclature is used:
YYMMDD_INSTRUMENT_NNN_FLOWCELL
where YYMMDD is the date, INSTRUMENT is the instrument name, NNN is the sequencing run number, and FLOWCELL is the flowcell ID.
NextSeq
NextSeq data is stored in /mnt/remoteserv/switch/rundata/nextseq/Runs and the instrument name is NB500898. Run ls to find out the last sequencing run.
ls -lrt | tail
drwxr-sr-x 8 tstuart listerlab 4096 Jan 28 10:33 180126_NB500898_031_HJ5MFBGX5
drwxr-sr-x 9 dvargas listerlab 4096 Feb 7 19:04 180207_NB500898_032_HF3CJBGX5
drwxr-sr-x 8 tstuart listerlab 4096 Feb 9 11:34 180208_NB500898_033_HF27VBGX5
drwxr-sr-x 8 tstuart listerlab 4096 Feb 11 12:33 180209_NB500898_034_HCYTNBGX5
drwxr-sr-x 8 tstuart listerlab 4096 Feb 20 10:18 180219_NB500898_035_HJ5LTBGX5
drwxrwsrwx 10 dvargas listerlab 4096 Mar 6 10:21 180223_NB500898_036_HJ5VGBGX5
drwxrwsrwx 9 jpflueger listerlab 4096 Mar 16 21:09 180315_NB500898_037_HJ575BGX5
drwxrwsrwx 13 jpflueger listerlab 4096 Apr 20 16:27 180418_NB500898_038_HJ5VTBGX5
drwxrwsrwx 9 dvargas listerlab 4096 May 16 00:39 180515_NB500898_039_HJ53NBGX5
drwxrwsrwx 8 dtang listerlab 4096 May 22 11:29 180521_NB500898_040_HJ5J5BGX5Since the last run was 040, our NNN will be 041.
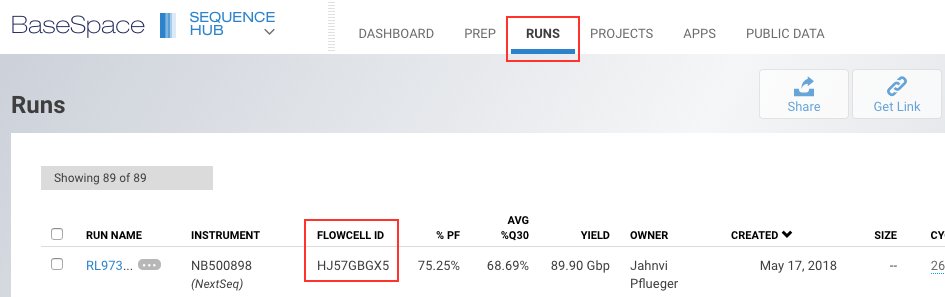
To get the flow cell information, we need to log into BaseSpace. Use the following credentials:
Email address: jahnvi.pflueger@uwa.edu.au
Password: (Ask Jahnvi for the password)
then go to the Dashboard and click on RUNS.
| Version | Author | Date |
|---|---|---|
| fd0493e | davetang | 2019-07-12 |
Now that we have all our information, we can create a new directory for our data. We will also make the directory fully accessible so that others can read and write to the directory.
cd /mnt/remoteserv/switch/rundata/nextseq/Runs
mkdir 180523_NB500898_041_HJ57GBGX5
chmod 777 180523_NB500898_041_HJ57GBGX5BaseMount
BaseMount is a tool to mount your BaseSpace Sequence Hub data as a Linux file system. Here’s the basic usage:
# Mount your BaseSpace account
mkdir BaseSpace
basemount BaseSpace/
<copy authentication URL to browser>
<login in browser>
<accept authentication>
# See the top level of your newly mounted environment!
ls BaseSpaceAuthentication
The first time you run BaseMount, you will be directed to a web URL and asked to enter your BaseSpace Sequence Hub user credentials. BaseMount will use these credentials to authenticate your interactions with BaseSpace Sequence Hub. By default, the credentials are cached in your home directory and they can be password-encrypted for security, just like an ssh key.
cat ~/.basespace/default.cfg
[DEFAULT]
accessToken = XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
apiServer = https://api.basespace.illumina.com/
[BaseMount]
tempDirectoryBaseName = /tmp/basemountThe next time you run BaseMount, you won’t need to perform the authentication step again.
Mounting
Now we can mount our BaseSpace Sequence Hub. You can call the directory anything you want but we’ll call it JahnviData.
cd /mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5
basemount JahnviData
,-----. ,--. ,--. ,--.
| |) /_ ,--,--. ,---. ,---. | `.' | ,---. ,--.,--.,--,--, ,-' '-.
| .-. \' ,-. |( .-' | .-. :| |'.'| || .-. || || || \'-. .-'
| '--' /\ '-' |.-' `)\ --.| | | |' '-' '' '' '| || | | |
`------' `--`--'`----' `----'`--' `--' `---' `----' `--''--' `--'
Illumina BaseMount v0.15.15.1872 public 2016-12-16 10:47
Command called:
basemount JahnviData
From:
/mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5
Mount point "JahnviData" doesn't exist
Create this mount point directory? (Y/n)
Creating directory "JahnviData"
Api Server: https://api.basespace.illumina.com/
Mounting BaseSpace account.
To unmount, run: basemount --unmount /mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5/JahnviDataIf we now go into JahnviData, we should see the following:
ls -lrt
total 1
drwxr-xr-x. 2 dtang dtang 0 May 23 12:30 Runs
-r--r--r--. 1 dtang dtang 598 May 23 12:30 README
drwxr-xr-x. 2 dtang dtang 0 May 23 12:30 ProjectsNext, find out the RUN NAME you want to download; the run name is available from the BaseSpace Sequence Hub RUN NAME column. In our case, the run name is RL973_Lister_2018_05_22, which is also the name of the directory. We want to go into the Files directory of our run.
cd /mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5/JahnviData/Runs/RL973_Lister_2018_05_22/Files
ls -lrt
total 57
-r--r--r--. 1 dtang dtang 26329 May 22 12:24 RunParameters.xml
-r--r--r--. 1 dtang dtang 28570 May 22 12:24 RunInfo.xml
-r--r--r--. 1 dtang dtang 37 May 22 20:42 RTARead1Complete.txt
-r--r--r--. 1 dtang dtang 37 May 22 21:34 RTARead2Complete.txt
-r--r--r--. 1 dtang dtang 37 May 23 11:18 RTARead3Complete.txt
-r--r--r--. 1 dtang dtang 47 May 23 11:18 RTAComplete.txt
-r--r--r--. 1 dtang dtang 926 May 23 11:36 RunCompletionStatus.xml
drwxr-xr-x. 2 dtang dtang 0 May 23 12:46 Thumbnail_Images
drwxr-xr-x. 2 dtang dtang 0 May 23 12:46 RTALogs
drwxr-xr-x. 2 dtang dtang 0 May 23 12:46 Logs
drwxr-xr-x. 2 dtang dtang 0 May 23 12:46 InterOp
drwxr-xr-x. 2 dtang dtang 0 May 23 12:46 DataNow we will use rsync to download the files locally; we use the parameters -ahPr --exclude Thumbnail_Images, which are:
- -a = archive mode
- -h = output numbers in a human-readable format
- -P = keep partially transferred files and show progress during transfer
- -r = recurse into directories
- –exclude Thumbnail_Images = exclude files matching Thumbnail_Images
Before you start the download, make sure you are in the right directory
pwd
/mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5/JahnviData/Runs/RL973_Lister_2018_05_22/Files
rsync -ahPr --exclude Thumbnail_Images * \
/dd_rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5 > \
/dd_rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5/copy.logThe copy.log file is used to ensure that each file is completely transferred.
head copy.log
sending incremental file list
RTAComplete.txt
47 100% 0.00kB/s 0:00:00 (xfer#1, to-check=1228/1229)
RTARead1Complete.txt
37 100% 0.03kB/s 0:00:01 (xfer#2, to-check=1227/1229)
RTARead2Complete.txt
37 100% 0.00kB/s 0:00:00 (xfer#3, to-check=1226/1229)
RTARead3Complete.txt
37 100% 0.03kB/s 0:00:01 (xfer#4, to-check=1225/1229)
RunCompletionStatus.xmlOnce downloading has completed, unmount the directory.
basemount --unmount /mnt/remoteserv/switch/rundata/nextseq/Runs/180523_NB500898_041_HJ57GBGX5/JahnviData
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.0.1 purrr_0.3.1
[5] readr_1.3.1 tidyr_0.8.3 tibble_2.0.1 ggplot2_3.1.0
[9] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 plyr_1.8.4 pillar_1.3.1
[5] compiler_3.5.2 git2r_0.24.0 workflowr_1.2.0 tools_3.5.2
[9] digest_0.6.18 lubridate_1.7.4 jsonlite_1.6 evaluate_0.13
[13] nlme_3.1-137 gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2
[17] rlang_0.3.1 cli_1.0.1 rstudioapi_0.9.0 yaml_2.2.0
[21] haven_2.1.0 xfun_0.5 withr_2.1.2 xml2_1.2.0
[25] httr_1.4.0 knitr_1.21 hms_0.4.2 generics_0.0.2
[29] fs_1.2.6 rprojroot_1.3-2 grid_3.5.2 tidyselect_0.2.5
[33] glue_1.3.0 R6_2.4.0 readxl_1.3.0 rmarkdown_1.11
[37] modelr_0.1.4 magrittr_1.5 whisker_0.3-2 backports_1.1.3
[41] scales_1.0.0 htmltools_0.3.6 rvest_0.3.2 assertthat_0.2.0
[45] colorspace_1.4-0 stringi_1.3.1 lazyeval_0.2.1 munsell_0.5.0
[49] broom_0.5.1 crayon_1.3.4