Using CloudStor
Dave Tang
2019-07-17
Last updated: 2019-07-17
Checks: 6 0
Knit directory: listerlab/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190712) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: analysis/.Rhistory
Untracked files:
Untracked: docker/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bb0a9d3 | davetang | 2019-07-17 | Progress bar |
| html | a0ef9b7 | davetang | 2019-07-12 | Build site. |
| Rmd | 389bd42 | davetang | 2019-07-12 | First commit |
CloudStor removes the frustration of slow data transfer rates for very large files by providing a super-fast, easy-to-use and secure file transfer and storage solution hosted on the AARNet network.
Unlike most cloud storage services, CloudStor is designed to meet the specific needs of researchers, and one terabyte of storage is available free to each individual researcher at AARNet-connected institutions.
Go to the URL https://www.aarnet.edu.au/network-and-services/cloud-services-applications/cloudstor and click “LOGIN TO CLOUDSTOR”, which should bring you to the UWA login page.
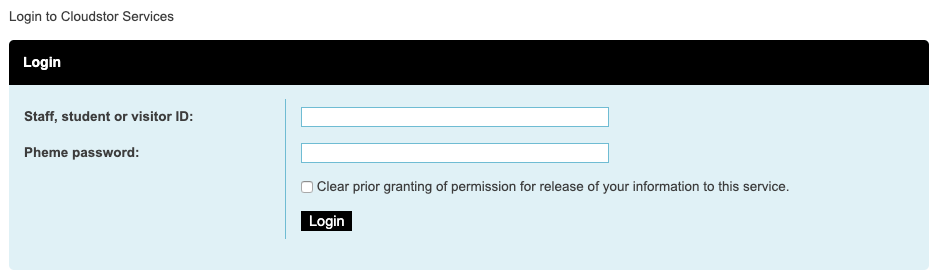
Once your username and password have been verified, you should land on your own CloudStor page.
Command line upload
You can manually upload files using the web interface but you can also use curl (CloudStor supports WebDAV), which makes it convenient for directly uploading files from a server to CloudStor.
curl --progress-bar --verbose -T my_file.txt --user dave.tang@uwa.edu.au https://cloudstor.aarnet.edu.au/plus/remote.php/webdav/my_dir/ | tee /dev/nullIn the example above, we are uploading my_file.txt to my_dir on CloudStor; change dave.tang@uwa.edu.au to your own UWA account. The command will only work if my_dir exists, so create it using the web interface prior to running the command. Otherwise, remove my_dir/ from the line and you can upload directly to the root directory. Do not forget the trailing /.
The command will prompt you for a password; use your CloudStor password that you created when you first logged onto CloudStor.
I’ve written a simple Perl script to help generate the command.
#!/usr/bin/env perl
use strict;
use warnings;
my $usage = "Usage: $0 <file> <dir>\n";
my $file = shift or die $usage;
my $dir = shift or die $usage;
my $user = "dave.tang\@uwa.edu.au";
my $command = "curl --progress-bar --verbose -T $file --user $user https://cloudstor.aarnet.edu.au/plus/remote.php/webdav/$dir/ | tee /dev/null";
print "Command to execute:\n\n$command\n\n";
print "Type yes to continue\n";
my $go = <STDIN>;
chomp($go);
if ($go =~ /^y/i){
system($command);
} else {
exit(0);
}
exit(0);You can also use curl to send other request methods, such as deleting a file.
curl -X DELETE --user dave.tang@uwa.edu.au https://cloudstor.aarnet.edu.au/plus/remote.php/webdav/my_file.txtDirectory upload
If you want to upload a directory, just use tar to create an archive first and then upload the tarball.
tar -czf my_dir.tar.gz my_dirFile sharing
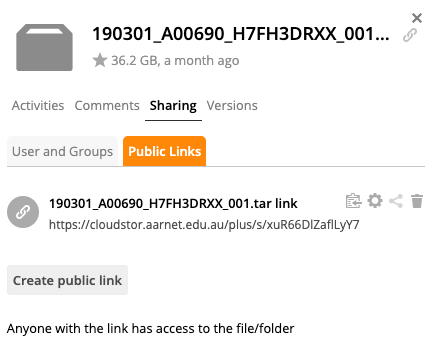
You can share files once you have successfully uploaded them. To share a file, click on the blank space directly after your file name (not the file icon or file text). Then click on “Sharing” and then “Public Links” and finally “Create public link”.
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.0.1 purrr_0.3.1
[5] readr_1.3.1 tidyr_0.8.3 tibble_2.0.1 ggplot2_3.1.0
[9] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 plyr_1.8.4 pillar_1.3.1
[5] compiler_3.5.2 git2r_0.24.0 workflowr_1.2.0 tools_3.5.2
[9] digest_0.6.18 lubridate_1.7.4 jsonlite_1.6 evaluate_0.13
[13] nlme_3.1-137 gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2
[17] rlang_0.3.1 cli_1.0.1 rstudioapi_0.9.0 yaml_2.2.0
[21] haven_2.1.0 xfun_0.5 withr_2.1.2 xml2_1.2.0
[25] httr_1.4.0 knitr_1.21 hms_0.4.2 generics_0.0.2
[29] fs_1.2.6 rprojroot_1.3-2 grid_3.5.2 tidyselect_0.2.5
[33] glue_1.3.0 R6_2.4.0 readxl_1.3.0 rmarkdown_1.11
[37] modelr_0.1.4 magrittr_1.5 whisker_0.3-2 backports_1.1.3
[41] scales_1.0.0 htmltools_0.3.6 rvest_0.3.2 assertthat_0.2.0
[45] colorspace_1.4-0 stringi_1.3.1 lazyeval_0.2.1 munsell_0.5.0
[49] broom_0.5.1 crayon_1.3.4