Seurat analysis using memory versus on disk
2025-03-05
Last updated: 2025-03-05
Checks: 7 0
Knit directory: muse/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200712) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 179e975. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: data/1M_neurons_filtered_gene_bc_matrices_h5.h5
Ignored: data/293t/
Ignored: data/293t_3t3_filtered_gene_bc_matrices.tar.gz
Ignored: data/293t_filtered_gene_bc_matrices.tar.gz
Ignored: data/5k_Human_Donor1_PBMC_3p_gem-x_5k_Human_Donor1_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor2_PBMC_3p_gem-x_5k_Human_Donor2_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor3_PBMC_3p_gem-x_5k_Human_Donor3_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor4_PBMC_3p_gem-x_5k_Human_Donor4_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/Parent_SC3v3_Human_Glioblastoma_filtered_feature_bc_matrix.tar.gz
Ignored: data/brain_counts/
Ignored: data/cl.obo
Ignored: data/cl.owl
Ignored: data/jurkat/
Ignored: data/jurkat:293t_50:50_filtered_gene_bc_matrices.tar.gz
Ignored: data/jurkat_293t/
Ignored: data/jurkat_filtered_gene_bc_matrices.tar.gz
Ignored: data/pbmc20k/
Ignored: data/pbmc20k_seurat/
Ignored: data/pbmc3k/
Ignored: data/pbmc4k_filtered_gene_bc_matrices.tar.gz
Ignored: data/pbmc_1k_v3_raw_feature_bc_matrix.h5
Ignored: data/refdata-gex-GRCh38-2020-A.tar.gz
Ignored: data/seurat_1m_neuron.rds
Ignored: data/t_3k_filtered_gene_bc_matrices.tar.gz
Ignored: r_packages_4.4.1/
Untracked files:
Untracked: analysis/bioc_scrnaseq.Rmd
Untracked: data/97516b79-8d08-46a6-b329-5d0a25b0be98.h5ad
Untracked: data/pbmc_1k_v3_filtered_feature_bc_matrix.h5
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/bpcells_vs_memory.Rmd) and
HTML (docs/bpcells_vs_memory.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 179e975 | Dave Tang | 2025-03-05 | Tabula Sapiens Heart cells |
| html | bc26143 | Dave Tang | 2025-02-24 | Build site. |
| Rmd | 8ead67f | Dave Tang | 2025-02-24 | Seurat memory vs on disk |
BPCells is an R package that allows for computationally efficient single-cell analysis. It utilizes bit-packing compression to store counts matrices on disk and C++ code to cache operations.
remotes::install_github("bnprks/BPCells/r")suppressPackageStartupMessages(library(BPCells))
suppressPackageStartupMessages(library(Seurat))PBMC3k
Download the Peripheral Blood Mononuclear Cells (PBMCs) 2,700 cells dataset.
mkdir -p data/pbmc3k && cd data/pbmc3k
wget -c https://s3-us-west-2.amazonaws.com/10x.files/samples/cell/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz
tar -xzf pbmc3k_filtered_gene_bc_matrices.tar.gzCreate Seurat object.
work_dir <- rprojroot::find_rstudio_root_file()
hdf5_file <- paste0(work_dir, "/data/5k_Human_Donor1_PBMC_3p_gem-x_5k_Human_Donor1_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5")
stopifnot(file.exists(hdf5_file))
seurat_obj_mem <- CreateSeuratObject(
counts = Seurat::Read10X_h5(hdf5_file),
min.cells = 3,
min.features = 200,
project = "pbmc3k"
)
class(seurat_obj_mem@assays$RNA$counts)[1] "dgCMatrix"
attr(,"package")
[1] "Matrix"Create Seurat object using {BPCells}.
seurat_obj_bpcells <- CreateSeuratObject(
counts = BPCells::open_matrix_10x_hdf5(hdf5_file),
min.cells = 3,
min.features = 200,
project = "pbmc3k"
)
seurat_obj_bpcells@assays$RNA$counts25348 x 5697 IterableMatrix object with class RenameDims
Row names: ENSG00000238009, ENSG00000239945 ... ENSG00000278817
Col names: AAACCAAAGGTGACGA-1, AAACCCTGTGACGAGT-1 ... TGTGTTGAGGATCTCA-1
Data type: uint32_t
Storage order: column major
Queued Operations:
1. 10x HDF5 feature matrix in file /home/rstudio/muse/data/5k_Human_Donor1_PBMC_3p_gem-x_5k_Human_Donor1_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
2. Select rows: 6, 7 ... 38605 and cols: 1, 2 ... 5710
3. Reset dimnamesSeurat workflow
Seurat version 4 workflow.
options(future.globals.maxSize = 1.5 * 1024^3)
fixed_PrepDR5 <- function(object, features = NULL, layer = 'scale.data', verbose = TRUE) {
layer <- layer[1L]
olayer <- layer
layer <- SeuratObject::Layers(object = object, search = layer)
if (is.null(layer)) {
abort(paste0("No layer matching pattern '", olayer, "' not found. Please run ScaleData and retry"))
}
data.use <- SeuratObject::LayerData(object = object, layer = layer)
features <- features %||% VariableFeatures(object = object)
if (!length(x = features)) {
stop("No variable features, run FindVariableFeatures() or provide a vector of features", call. = FALSE)
}
if (is(data.use, "IterableMatrix")) {
features.var <- BPCells::matrix_stats(matrix=data.use, row_stats="variance")$row_stats["variance",]
} else {
features.var <- apply(X = data.use, MARGIN = 1L, FUN = var)
}
features.keep <- features[features.var > 0]
if (!length(x = features.keep)) {
stop("None of the requested features have any variance", call. = FALSE)
} else if (length(x = features.keep) < length(x = features)) {
exclude <- setdiff(x = features, y = features.keep)
if (isTRUE(x = verbose)) {
warning(
"The following ",
length(x = exclude),
" features requested have zero variance; running reduction without them: ",
paste(exclude, collapse = ', '),
call. = FALSE,
immediate. = TRUE
)
}
}
features <- features.keep
features <- features[!is.na(x = features)]
features.use <- features[features %in% rownames(data.use)]
if(!isTRUE(all.equal(features, features.use))) {
missing_features <- setdiff(features, features.use)
if(length(missing_features) > 0) {
warning_message <- paste("The following features were not available: ",
paste(missing_features, collapse = ", "),
".", sep = "")
warning(warning_message, immediate. = TRUE)
}
}
data.use <- data.use[features.use, ]
return(data.use)
}
assignInNamespace('PrepDR5', fixed_PrepDR5, 'Seurat')
seurat_workflow <- function(obj, debug_flag = FALSE){
obj <- NormalizeData(obj, normalization.method = "LogNormalize", verbose = debug_flag)
obj <- FindVariableFeatures(obj, selection.method = 'vst', nfeatures = 2000, verbose = debug_flag)
obj <- ScaleData(obj, verbose = debug_flag)
obj <- RunPCA(obj, verbose = debug_flag)
obj <- RunUMAP(obj, dims = 1:30, verbose = debug_flag)
obj <- FindNeighbors(obj, dims = 1:30, verbose = debug_flag)
obj <- FindClusters(obj, resolution = 0.5, verbose = debug_flag)
obj
}Run workflow for in memory.
start_time <- Sys.time()
seurat_obj_mem <- seurat_workflow(seurat_obj_mem)Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per sessionend_time <- Sys.time()
end_time - start_timeTime difference of 15.38153 secsRun workflow for on disk.
start_time <- Sys.time()
seurat_obj_bpcells <- seurat_workflow(seurat_obj_bpcells)
end_time <- Sys.time()
end_time - start_timeTime difference of 3.914375 minsResults
Compare clustering.
stopifnot(all(row.names(seurat_obj_bpcells@meta.data) == row.names(seurat_obj_mem@meta.data)))
table(
seurat_obj_bpcells@meta.data$seurat_clusters,
seurat_obj_mem@meta.data$seurat_clusters
)
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
0 1069 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 0 769 0 0 0 0 0 0 0 0 0 0 0 0 0
2 0 0 701 0 0 0 0 0 0 0 0 0 0 0 0
3 0 0 0 527 0 0 0 0 0 0 0 0 0 0 0
4 0 0 0 0 476 0 0 0 0 0 0 0 0 0 0
5 0 0 0 0 0 453 0 0 0 0 0 0 0 0 0
6 0 0 0 0 0 0 387 0 0 0 0 0 0 0 0
7 0 0 0 0 0 0 0 380 0 0 0 0 0 0 0
8 0 0 0 0 0 0 0 0 336 0 0 0 0 0 0
9 0 0 0 0 0 0 0 0 0 249 0 0 0 0 0
10 0 0 0 0 0 0 0 0 0 0 158 0 0 0 0
11 0 0 0 0 0 0 0 0 0 0 0 90 0 0 0
12 0 0 0 0 0 0 0 0 0 0 0 0 52 0 0
13 0 0 0 0 0 0 0 0 0 0 0 0 0 34 0
14 0 0 0 0 0 0 0 0 0 0 0 0 0 0 16Tabula Sapiens
Download from Tabula Sapiens:
Tabula Sapiens is a benchmark, first-draft human cell atlas of over 1.1M cells from 28 organs of 24 normal human subjects. This work is the product of the Tabula Sapiens Consortium. Taking the organs from the same individual controls for genetic background, age, environment, and epigenetic effects, and allows detailed analysis and comparison of cell types that are shared between tissues.
ts_heart_url <- "https://datasets.cellxgene.cziscience.com/97516b79-8d08-46a6-b329-5d0a25b0be98.h5ad"
ts_heart_file <- paste0('data/', basename(ts_heart_url))
if(file.exists(ts_heart_file)){
message(paste0(ts_heart_file, " already exists"))
} else {
options(timeout = 1000)
download.file(url = ts_heart_url, destfile = ts_heart_file)
}data/97516b79-8d08-46a6-b329-5d0a25b0be98.h5ad already existsRun workflow for on disk.
start_time <- Sys.time()
ts_heart <- CreateSeuratObject(
counts = BPCells::open_matrix_anndata_hdf5(ts_heart_file),
min.cells = 3,
min.features = 200,
project = "ts_heart"
)
ts_heart <- seurat_workflow(ts_heart)
end_time <- Sys.time()
end_time - start_timeTime difference of 23.4599 minsUMAP.
DimPlot(ts_heart)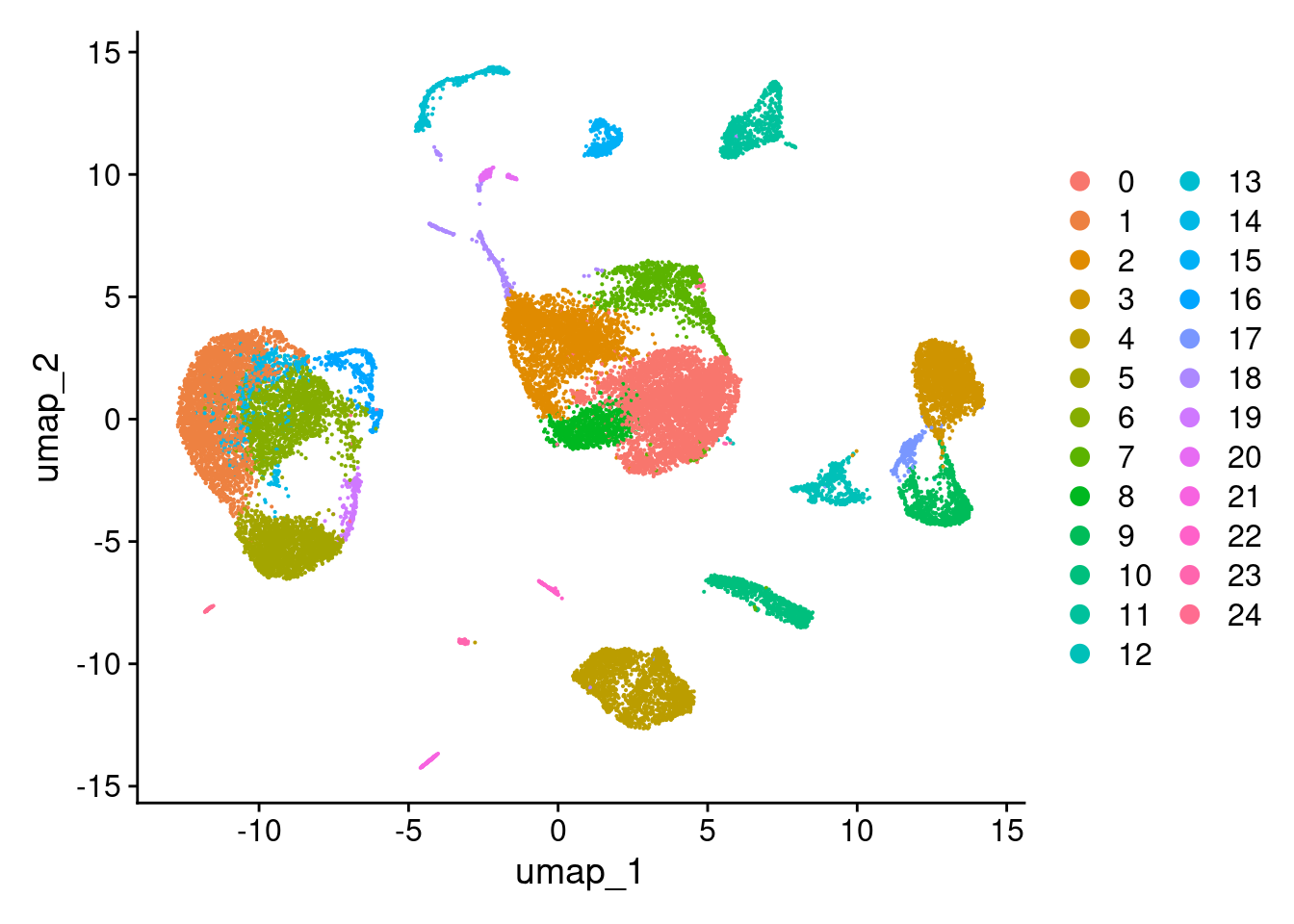
sessionInfo()R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] Seurat_5.1.0 SeuratObject_5.0.2 sp_2.1-4 BPCells_0.3.0
[5] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 rstudioapi_0.17.1 jsonlite_1.8.9
[4] magrittr_2.0.3 spatstat.utils_3.1-0 farver_2.1.2
[7] rmarkdown_2.28 fs_1.6.4 vctrs_0.6.5
[10] ROCR_1.0-11 spatstat.explore_3.3-3 htmltools_0.5.8.1
[13] sass_0.4.9 sctransform_0.4.1 parallelly_1.38.0
[16] KernSmooth_2.23-24 bslib_0.8.0 htmlwidgets_1.6.4
[19] ica_1.0-3 plyr_1.8.9 plotly_4.10.4
[22] zoo_1.8-12 cachem_1.1.0 whisker_0.4.1
[25] igraph_2.1.1 mime_0.12 lifecycle_1.0.4
[28] pkgconfig_2.0.3 Matrix_1.7-0 R6_2.5.1
[31] fastmap_1.2.0 MatrixGenerics_1.18.1 fitdistrplus_1.2-1
[34] future_1.34.0 shiny_1.9.1 digest_0.6.37
[37] colorspace_2.1-1 patchwork_1.3.0 ps_1.8.1
[40] rprojroot_2.0.4 tensor_1.5 RSpectra_0.16-2
[43] irlba_2.3.5.1 labeling_0.4.3 progressr_0.15.0
[46] fansi_1.0.6 spatstat.sparse_3.1-0 httr_1.4.7
[49] polyclip_1.10-7 abind_1.4-8 compiler_4.4.1
[52] withr_3.0.2 bit64_4.5.2 fastDummies_1.7.4
[55] highr_0.11 MASS_7.3-60.2 tools_4.4.1
[58] lmtest_0.9-40 httpuv_1.6.15 future.apply_1.11.3
[61] goftest_1.2-3 glue_1.8.0 callr_3.7.6
[64] nlme_3.1-164 promises_1.3.0 grid_4.4.1
[67] Rtsne_0.17 getPass_0.2-4 cluster_2.1.6
[70] reshape2_1.4.4 generics_0.1.3 hdf5r_1.3.11
[73] gtable_0.3.6 spatstat.data_3.1-2 tidyr_1.3.1
[76] data.table_1.16.2 utf8_1.2.4 spatstat.geom_3.3-3
[79] RcppAnnoy_0.0.22 ggrepel_0.9.6 RANN_2.6.2
[82] pillar_1.9.0 stringr_1.5.1 spam_2.11-0
[85] RcppHNSW_0.6.0 later_1.3.2 splines_4.4.1
[88] dplyr_1.1.4 lattice_0.22-6 bit_4.5.0
[91] survival_3.6-4 deldir_2.0-4 tidyselect_1.2.1
[94] miniUI_0.1.1.1 pbapply_1.7-2 knitr_1.48
[97] git2r_0.35.0 gridExtra_2.3 scattermore_1.2
[100] xfun_0.48 matrixStats_1.4.1 stringi_1.8.4
[103] lazyeval_0.2.2 yaml_2.3.10 evaluate_1.0.1
[106] codetools_0.2-20 tibble_3.2.1 cli_3.6.3
[109] uwot_0.2.2 xtable_1.8-4 reticulate_1.39.0
[112] munsell_0.5.1 processx_3.8.4 jquerylib_0.1.4
[115] Rcpp_1.0.13 globals_0.16.3 spatstat.random_3.3-2
[118] png_0.1-8 spatstat.univar_3.0-1 parallel_4.4.1
[121] ggplot2_3.5.1 dotCall64_1.2 listenv_0.9.1
[124] viridisLite_0.4.2 scales_1.3.0 ggridges_0.5.6
[127] leiden_0.4.3.1 purrr_1.0.2 rlang_1.1.4
[130] cowplot_1.1.3