Creating a Seurat object
2025-02-17
Last updated: 2025-02-17
Checks: 7 0
Knit directory: muse/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200712) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 58b172f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: data/1M_neurons_filtered_gene_bc_matrices_h5.h5
Ignored: data/293t/
Ignored: data/293t_3t3_filtered_gene_bc_matrices.tar.gz
Ignored: data/293t_filtered_gene_bc_matrices.tar.gz
Ignored: data/5k_Human_Donor1_PBMC_3p_gem-x_5k_Human_Donor1_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor2_PBMC_3p_gem-x_5k_Human_Donor2_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor3_PBMC_3p_gem-x_5k_Human_Donor3_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/5k_Human_Donor4_PBMC_3p_gem-x_5k_Human_Donor4_PBMC_3p_gem-x_count_sample_filtered_feature_bc_matrix.h5
Ignored: data/Parent_SC3v3_Human_Glioblastoma_filtered_feature_bc_matrix.tar.gz
Ignored: data/brain_counts/
Ignored: data/cl.obo
Ignored: data/cl.owl
Ignored: data/jurkat/
Ignored: data/jurkat:293t_50:50_filtered_gene_bc_matrices.tar.gz
Ignored: data/jurkat_293t/
Ignored: data/jurkat_filtered_gene_bc_matrices.tar.gz
Ignored: data/pbmc4k_filtered_gene_bc_matrices.tar.gz
Ignored: data/refdata-gex-GRCh38-2020-A.tar.gz
Ignored: data/seurat_1m_neuron.rds
Ignored: data/t_3k_filtered_gene_bc_matrices.tar.gz
Ignored: r_packages_4.4.1/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/seurat_object.Rmd) and
HTML (docs/seurat_object.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 58b172f | Dave Tang | 2025-02-17 | Join layers |
| html | 30a9f5e | Dave Tang | 2025-02-17 | Build site. |
| Rmd | bef6e98 | Dave Tang | 2025-02-17 | Seurat integration |
| html | 5e87899 | Dave Tang | 2025-02-17 | Build site. |
| Rmd | 1f44cd2 | Dave Tang | 2025-02-17 | Creating a Seurat object |
Load {Seurat} and {hdf5r}.
suppressPackageStartupMessages(library("Seurat"))
suppressPackageStartupMessages(library("hdf5r"))
packageVersion("Seurat")[1] '5.1.0'Data
Read HDF5 files into a list.
hdf5_files <- list.files(path = "data", pattern = "5k_Human", full.names = TRUE)
mats <- purrr::map(seq_along(hdf5_files), function(x){
my_mat <- Seurat::Read10X_h5(hdf5_files[x])
colnames(my_mat) <- paste0('donor', x, '_', colnames(my_mat))
my_mat
})
str(mats, max.level = 1)List of 4
$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
$ :Formal class 'dgCMatrix' [package "Matrix"] with 6 slotsCreate Seurat object using a list of matrices.
seurat_object <- CreateSeuratObject(
counts = mats,
min.cells = 3,
min.features = 200
)
seurat_objectAn object of class Seurat
27385 features across 22061 samples within 1 assay
Active assay: RNA (27385 features, 0 variable features)
4 layers present: counts.1, counts.2, counts.3, counts.4Somehow orig.ident now has the donor information.
seurat_object@meta.data |>
tail() orig.ident nCount_RNA nFeature_RNA
donor4_TGTGGTTGTGCGCAAG-1 donor4 13490 3701
donor4_TGTGGTTGTTCGCACA-1 donor4 4686 1683
donor4_TGTGGTTGTTGAGGCG-1 donor4 8545 2748
donor4_TGTGTACGTCAGCATT-1 donor4 8910 3003
donor4_TGTGTTGAGATTGCTG-1 donor4 14597 3977
donor4_TGTGTTGAGTTACGGC-1 donor4 12983 3316Merging
Create one count layer.
seurat_object_joined <- JoinLayers(seurat_object)
seurat_object_joinedAn object of class Seurat
27385 features across 22061 samples within 1 assay
Active assay: RNA (27385 features, 0 variable features)
1 layer present: countsTail of metadata.
seurat_object_joined@meta.data |>
tail() orig.ident nCount_RNA nFeature_RNA
donor4_TGTGGTTGTGCGCAAG-1 donor4 13490 3701
donor4_TGTGGTTGTTCGCACA-1 donor4 4686 1683
donor4_TGTGGTTGTTGAGGCG-1 donor4 8545 2748
donor4_TGTGTACGTCAGCATT-1 donor4 8910 3003
donor4_TGTGTTGAGATTGCTG-1 donor4 14597 3977
donor4_TGTGTTGAGTTACGGC-1 donor4 12983 3316Integration
Integrative analysis in Seurat v5:
Integration of single-cell sequencing datasets, for example across experimental batches, donors, or conditions, is often an important step in scRNA-seq workflows. Integrative analysis can help to match shared cell types and states across datasets, which can boost statistical power, and most importantly, facilitate accurate comparative analysis across datasets.
Process.
verbose_mode <- FALSE
seurat_object <- NormalizeData(seurat_object, verbose = verbose_mode)
seurat_object <- FindVariableFeatures(seurat_object, verbose = verbose_mode)
seurat_object <- ScaleData(seurat_object, verbose = verbose_mode)
seurat_object <- RunPCA(seurat_object, verbose = verbose_mode)
seurat_object <- FindNeighbors(seurat_object, dims = 1:30, reduction = "pca", verbose = verbose_mode)
seurat_object <- FindClusters(seurat_object, resolution = 0.5, cluster.name = "unintegrated_clusters", verbose = verbose_mode)
seurat_object <- RunUMAP(seurat_object, dims = 1:30, reduction = "pca", reduction.name = "umap.unintegrated", verbose = verbose_mode)Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per sessionDimPlot(seurat_object, reduction = "umap.unintegrated", group.by = "orig.ident")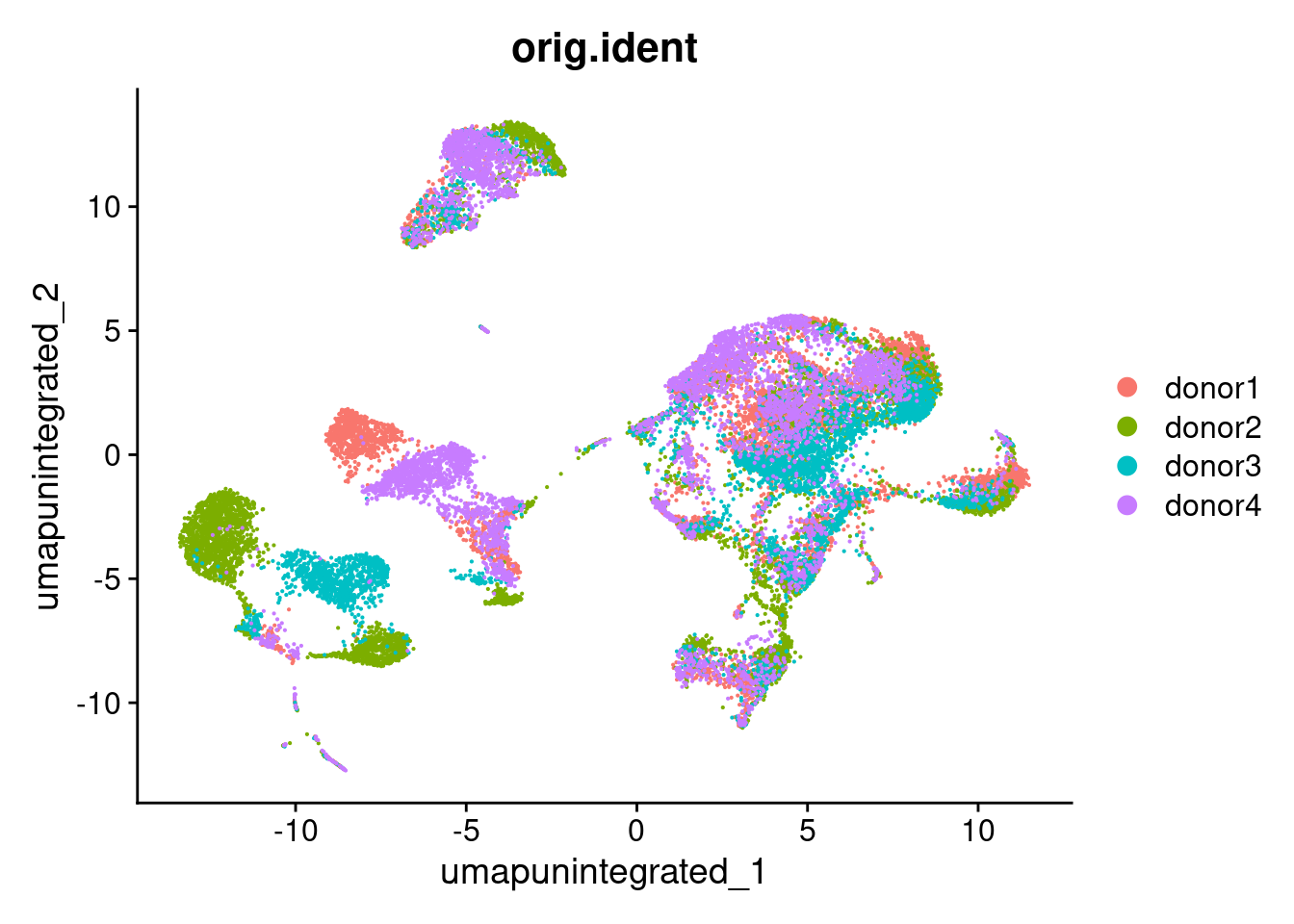
| Version | Author | Date |
|---|---|---|
| 30a9f5e | Dave Tang | 2025-02-17 |
Harmony integration.
seurat_object <- IntegrateLayers(
object = seurat_object,
method = HarmonyIntegration,
orig.reduction = "pca",
new.reduction = "harmony",
verbose = verbose_mode
)Warning in harmony::HarmonyMatrix(data_mat = Embeddings(object = orig), :
HarmonyMatrix is deprecated and will be removed in the future from the API in
the futureWarning: Warning: The parameters do_pca and npcs are deprecated. They will be ignored for this function call and please remove parameters do_pca and npcs and pass to harmony cell_embeddings directly.
This warning is displayed once per session.Warning: Warning: The parameter tau is deprecated. It will be ignored for this function call and please remove parameter tau in future function calls. Advanced users can set value of parameter tau by using parameter .options and function harmony_options().
This warning is displayed once per session.Warning: Warning: The parameter block.size is deprecated. It will be ignored for this function call and please remove parameter block.size in future function calls. Advanced users can set value of parameter block.size by using parameter .options and function harmony_options().
This warning is displayed once per session.Warning: Warning: The parameter max.iter.harmony is replaced with parameter max_iter. It will be ignored for this function call and please use parameter max_iter in future function calls.
This warning is displayed once per session.Warning: Warning: The parameter max.iter.cluster is deprecated. It will be ignored for this function call and please remove parameter max.iter.cluster in future function calls. Advanced users can set value of parameter max.iter.cluster by using parameter .options and function harmony_options().
This warning is displayed once per session.Warning: Warning: The parameter epsilon.cluster is deprecated. It will be ignored for this function call and please remove parameter epsilon.cluster in future function calls. Advanced users can set value of parameter epsilon.cluster by using parameter .options and function harmony_options().
This warning is displayed once per session.Warning: Warning: The parameter epsilon.harmony is deprecated. It will be ignored for this function call and please remove parameter epsilon.harmony in future function calls. If users want to control if harmony would stop early or not, use parameter early_stop. Advanced users can set value of parameter epsilon.harmony by using parameter .options and function harmony_options().
This warning is displayed once per session.seurat_object <- FindNeighbors(seurat_object, reduction = "harmony", dims = 1:30, verbose = verbose_mode)
seurat_object <- FindClusters(seurat_object, resolution = 0.5, cluster.name = "harmony_clusters", verbose = verbose_mode)
seurat_object <- RunUMAP(seurat_object, reduction = "harmony", dims = 1:30, reduction.name = "umap.harmony", verbose = verbose_mode)
DimPlot(seurat_object, reduction = "umap.harmony", group.by = "orig.ident")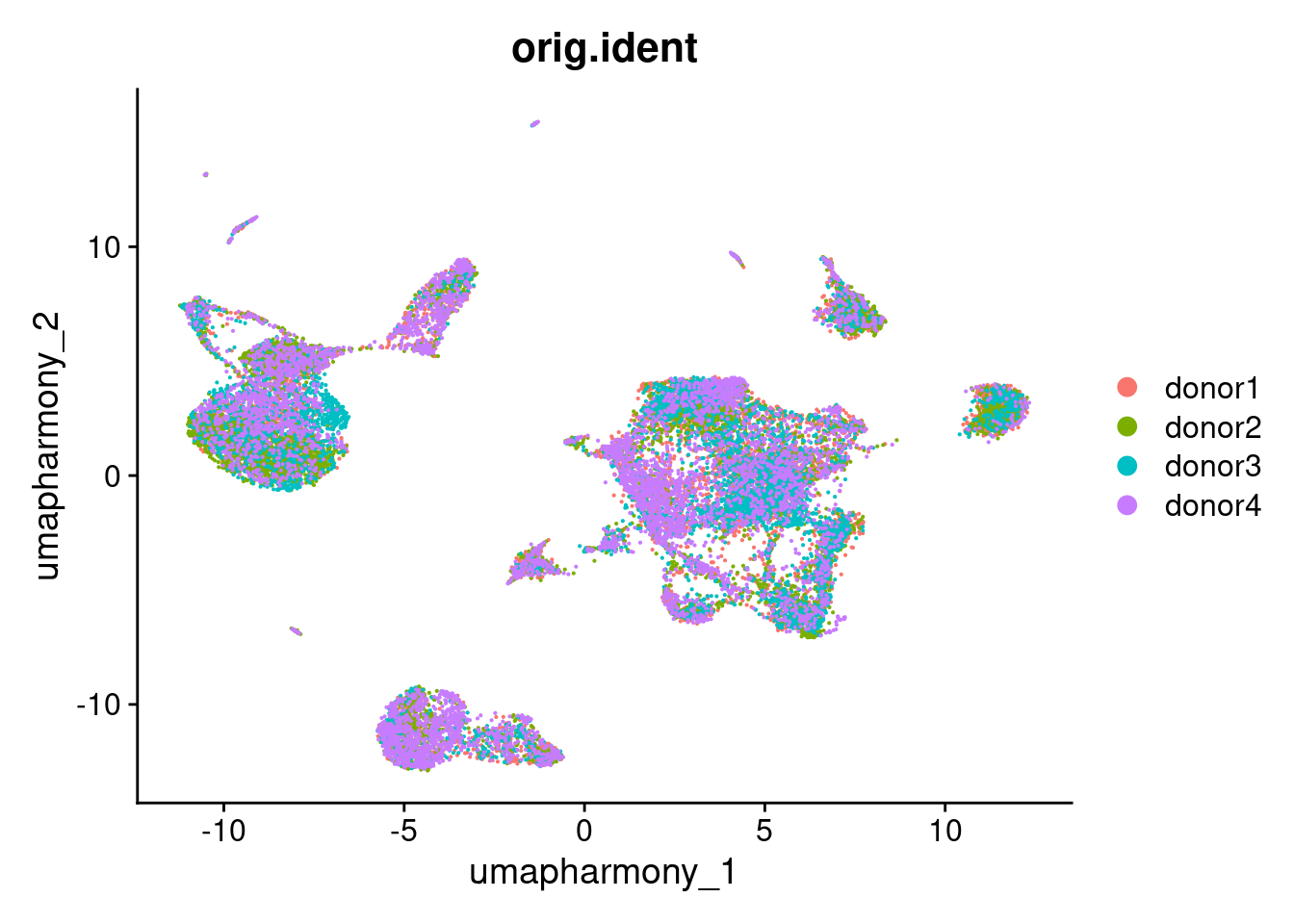
| Version | Author | Date |
|---|---|---|
| 30a9f5e | Dave Tang | 2025-02-17 |
sessionInfo()R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] hdf5r_1.3.11 Seurat_5.1.0 SeuratObject_5.0.2 sp_2.1-4
[5] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 rstudioapi_0.17.1 jsonlite_1.8.9
[4] magrittr_2.0.3 spatstat.utils_3.1-0 farver_2.1.2
[7] rmarkdown_2.28 fs_1.6.4 vctrs_0.6.5
[10] ROCR_1.0-11 spatstat.explore_3.3-3 htmltools_0.5.8.1
[13] sass_0.4.9 sctransform_0.4.1 parallelly_1.38.0
[16] KernSmooth_2.23-24 bslib_0.8.0 htmlwidgets_1.6.4
[19] ica_1.0-3 plyr_1.8.9 plotly_4.10.4
[22] zoo_1.8-12 cachem_1.1.0 whisker_0.4.1
[25] igraph_2.1.1 mime_0.12 lifecycle_1.0.4
[28] pkgconfig_2.0.3 Matrix_1.7-0 R6_2.5.1
[31] fastmap_1.2.0 fitdistrplus_1.2-1 future_1.34.0
[34] shiny_1.9.1 digest_0.6.37 colorspace_2.1-1
[37] patchwork_1.3.0 ps_1.8.1 rprojroot_2.0.4
[40] tensor_1.5 RSpectra_0.16-2 irlba_2.3.5.1
[43] labeling_0.4.3 progressr_0.15.0 fansi_1.0.6
[46] spatstat.sparse_3.1-0 httr_1.4.7 polyclip_1.10-7
[49] abind_1.4-8 compiler_4.4.1 withr_3.0.2
[52] bit64_4.5.2 fastDummies_1.7.4 highr_0.11
[55] MASS_7.3-60.2 tools_4.4.1 lmtest_0.9-40
[58] httpuv_1.6.15 future.apply_1.11.3 goftest_1.2-3
[61] glue_1.8.0 callr_3.7.6 nlme_3.1-164
[64] promises_1.3.0 grid_4.4.1 Rtsne_0.17
[67] getPass_0.2-4 cluster_2.1.6 reshape2_1.4.4
[70] generics_0.1.3 gtable_0.3.6 spatstat.data_3.1-2
[73] tidyr_1.3.1 data.table_1.16.2 utf8_1.2.4
[76] spatstat.geom_3.3-3 RcppAnnoy_0.0.22 ggrepel_0.9.6
[79] RANN_2.6.2 pillar_1.9.0 stringr_1.5.1
[82] spam_2.11-0 RcppHNSW_0.6.0 later_1.3.2
[85] splines_4.4.1 dplyr_1.1.4 lattice_0.22-6
[88] bit_4.5.0 survival_3.6-4 deldir_2.0-4
[91] tidyselect_1.2.1 miniUI_0.1.1.1 pbapply_1.7-2
[94] knitr_1.48 git2r_0.35.0 gridExtra_2.3
[97] scattermore_1.2 RhpcBLASctl_0.23-42 xfun_0.48
[100] matrixStats_1.4.1 stringi_1.8.4 lazyeval_0.2.2
[103] yaml_2.3.10 evaluate_1.0.1 codetools_0.2-20
[106] tibble_3.2.1 cli_3.6.3 uwot_0.2.2
[109] xtable_1.8-4 reticulate_1.39.0 munsell_0.5.1
[112] processx_3.8.4 jquerylib_0.1.4 harmony_1.2.1
[115] Rcpp_1.0.13 globals_0.16.3 spatstat.random_3.3-2
[118] png_0.1-8 spatstat.univar_3.0-1 parallel_4.4.1
[121] ggplot2_3.5.1 dotCall64_1.2 listenv_0.9.1
[124] viridisLite_0.4.2 scales_1.3.0 ggridges_0.5.6
[127] leiden_0.4.3.1 purrr_1.0.2 rlang_1.1.4
[130] cowplot_1.1.3